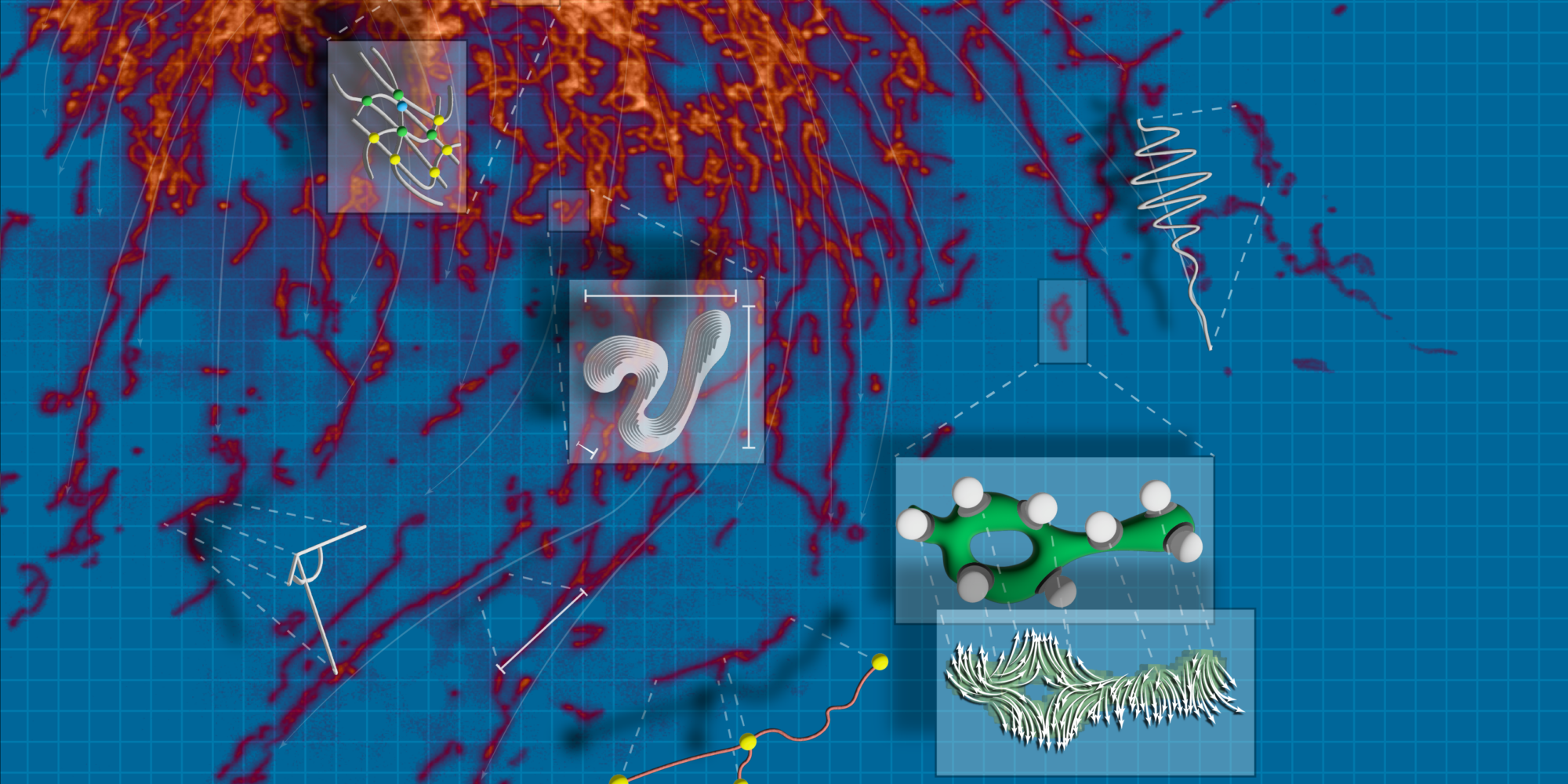
Scientists studying aging and disease have long been interested in organelles, the tiny structures inside cells that perform essential functions such as producing energy, processing and transporting proteins, and breaking down waste. However, their microscopic size and constant movement make them challenging to track. Advances in 2D and 3D imaging have provided clearer views of organelles in action, but the ability to extract meaningful insights from these images has been limited by the lack of automated, adaptable analysis tools.
To address these challenges, scientists at Calico have developed an automated image analysis tool that is helping researchers around the world learn more about organelles. The freely available algorithm, known as Nellie, was designed to perform segmentation, tracking, and feature extraction using 2D or 3D live-cell microscopy images, making these advanced analytical techniques accessible to more researchers.
Nellie was born out of the diverse imaging challenges at Calico, where scientists use a range of microscopy techniques to study organelles across different cell types. Understanding how organelles function in young versus aged cells, for example, can provide crucial insights into the earliest cellular changes associated with aging. Recognizing the need for a unified, adaptable solution, Senior Data Scientist Austin Lefebvre combined key elements from various specialized tools he had previously developed, each designed to track specific organelles based on their size, shape, and movement patterns. The result was Nellie, a single, comprehensive pipeline capable of analyzing virtually any organelle across multiple imaging types.
Nellie, which was released online last year, has now been reported in Nature Methods. In an extensive review of the algorithm, Calico scientists tested Nellie’s capabilities through three challenging applications: unmixing multiple types of organelles from a single fluorescence channel, analyzing drug-induced mitochondrial network changes, and characterizing endoplasmic reticulum dynamics. The tool successfully analyzed diverse organelle types without parameter tuning, exceeding the accuracy of specialized solutions.
“Most existing solutions demand extensive parameter tuning or expertise with deep-learning models, creating barriers for many researchers,” said Austin. “With Nellie, all you do is download one thing, and everything else is point and click. It’s very intuitive, while also being very versatile.” Since Nellie was posted to Github a year ago, it has been downloaded more than 900 times.
Future development of Nellie will focus on integrating optional deep-learning capabilities, enhancing tools for studying organelle interactions, and applying these features to large-scale phenotyping. The team also aims to expand Nellie’s functionality to analyze multiple organelles simultaneously, providing a more comprehensive view of cellular dynamics. Ultimately, the goal for Nellie is to serve as a catalyst for a new wave of scientific inquiry, where the complex weave and elaborate dance of organelles is not just observed, but deeply understood, and where the mysteries of the cell are unlocked, one pixel at a time.
Learn more about this tool in the Nature Methods publication, “Nellie: automated organelle segmentation, tracking and hierarchical feature extraction in 2D/3D live-cell microscopy.” To access the freely available Nellie pipeline and its Napari-based plugin online visit GitHub at https://github.com/aelefebv/nellie.